Research
Bacterial protein toxins are sophisticated protein nanomachines honed by evolution. They play central roles in bacterial pathogenesis and are responsible for many deadly infectious diseases. Once released from bacteria, these nanomachines act autonomously as “guided missiles”: targeting cells by binding to receptors, some acting directly on cell surfaces, and the most potent ones crossing the membrane into the cytosol and acting on key nodes in cellular processes.
The beauty of toxins lies in their specificity of action. On one hand, understanding and disarming toxins has led to prevention and treatment of many infectious diseases. On the other hand, a mechanistic understanding of toxin action has provided pivotal insights and powerful tools to pinpoint some of the most important players in complex cellular processes. The multiple steps of toxin action are efficiently achieved by multi-domain proteins that have been sculpted by evolution. Fundamentally, we believe these nanomachines serve as excellent models from which to discover the principles governing the behavior of proteins on receptor-recognition, membrane translocation, intracellular trafficking, and post-translational modifications.
The central goals of our lab include understanding the mechanisms of bacterial toxins, developing therapeutic approaches against toxins, and engineering toxin-inspired tools and therapeutics. Our laboratory has a broad interest in microbial toxins, bacterial pathogenesis, microbial interactions with human/animals/insects, microbiome, and engineering therapeutic proteins. Along these lines of basic research, we are keen in developing microbial protein-inspired novel therapeutics for treating genetic diseases, cancer, pain, and other neurological disorders. We have a multi-disciplinary team with diverse expertise in microbiology, structural biology, protein engineering, cell biology, insect biology, and neuroscience fields.
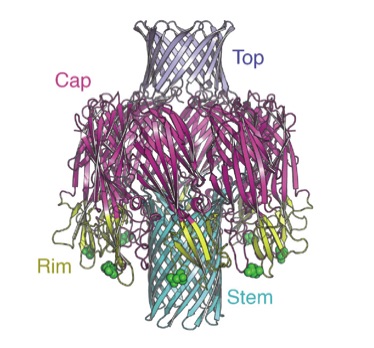
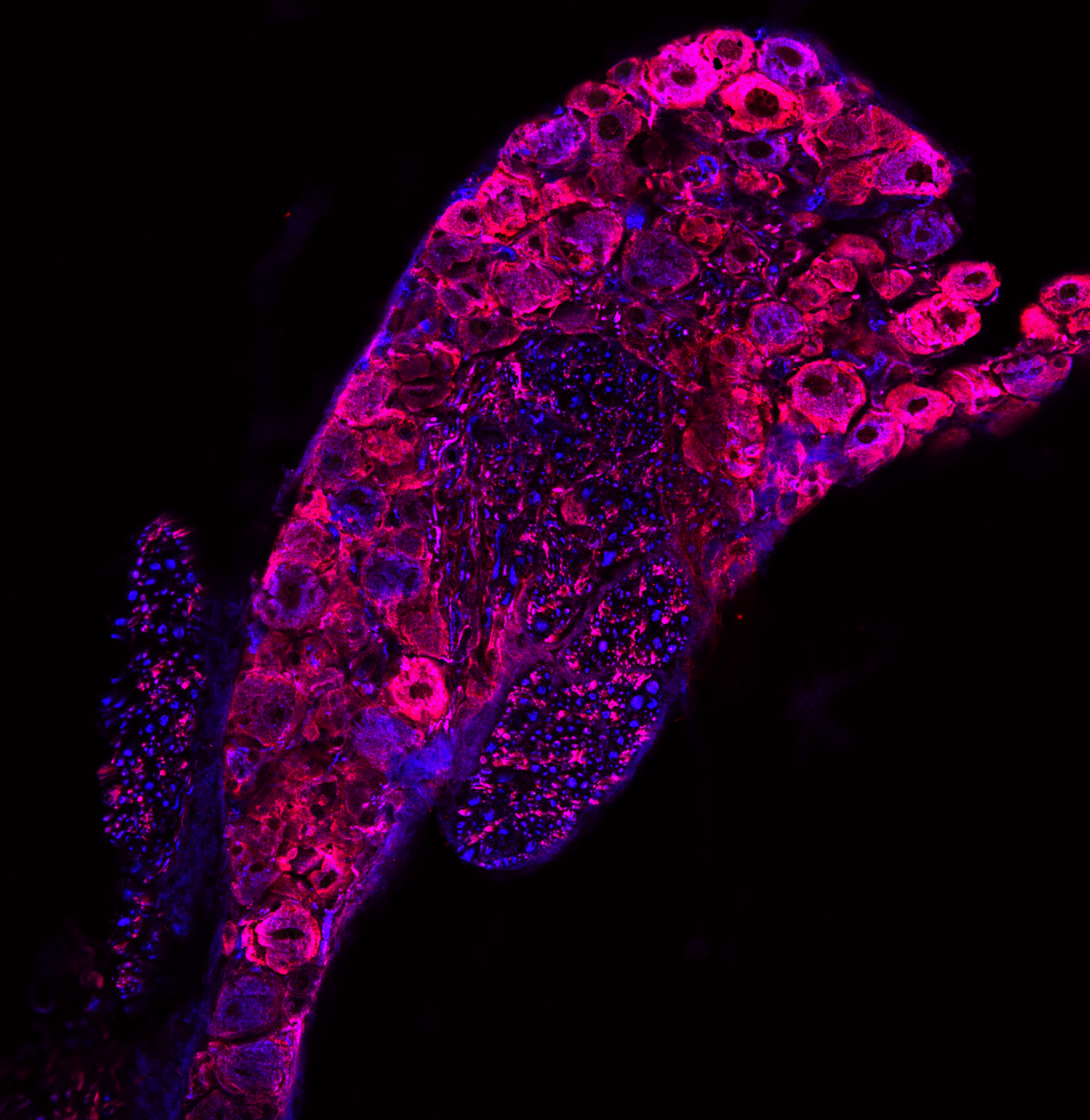
Pore structure of pore-forming toxin Epx4
Our research covers major bacterial toxins including botulinum neurotoxins (BoNTs, botox), tetanus neurotoxin, C. difficile toxins (TcdA and TcdB), Shiga toxins, insecticidal Tc toxins, and a growing number of novel microbial protein toxins with completely unknown biology. Our research mainly focused on identification of their host factors (e.g. receptors, trafficking factors, enzymatic targets) and understanding how these toxins modulate host physiology/immunity/pathogenesis at structural and functional levels.
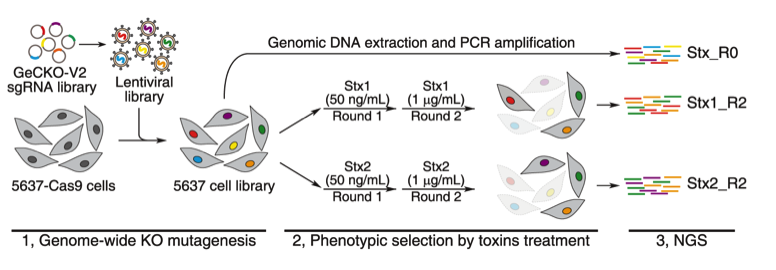
Recent representative achievements include identification of receptors for C. difficile toxin via genome-wide CRISPR-Cas9 screen (Nature, 2016, 538:350, Science, 2018, 360:664; Cell Host and Microbe, 2020, 27:782), identification and characterization of novel toxins (Nature Communications, 2017, 8:14130; Cell Host and Microbe, 2018, 23:169, Cell, 2022, 185:1), and protein engineering of botulinum neurotoxins (Nature Communications, 2017, 8:53; Science Translational Medicine, 2021, 13:eaaz4197; Science, 2021, 371:803).
Ongoing projects are in the following areas:
(1) Molecular and structural mechanisms of bacterial toxins, bacterial infections, and host-pathogen and microbiome-pathogen interactions;
(2) Engineering therapeutic proteins for genetic editing and modulation of signaling networks in cells for treating genetic diseases, pain, neurological disorders, and cancer;
(3) Developing therapeutic antibodies and vaccines against emerging pathogens;
(4) Neuron-immune interactions/regulations and membrane trafficking in neurons.
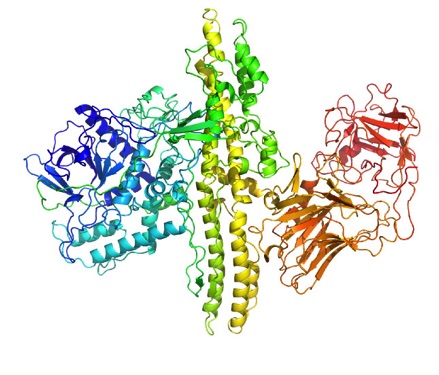
Protein Structure of Botulinum neurotoxin Type A
Protein Structure of Shiga toxin type 2
CRISPR-Cas9 screening for Shiga toxin receptor